Value of TMEM115 as a Potential Prognostic Biomarker in Liver Hepatocellular Carcinoma
Article information
Abstract
Transmembrane protein 115 (TMEM115) is a membrane protein; considering the potential of membrane proteins as biomarkers for various pathological conditions, we aimed to examine the value of TMEM115 as a potential biomarker for improving the prognosis and treatment of liver hepatocellular carcinoma (LIHC) in this study. Online databases including the Tumor Immune Estimation Resource, UALCAN, Gene Expression Profiling Interactive Analysis version 2, OSlihc, and human Protein Atlas were used. The analysis suggested that TMEM115 expression in LIHC was higher compared to normal tissues; further, TMEM115 expression was confirmed to be related with poor prognosis in LIHC. Higher protein expression levels of TMEM115 were observed in LIHC tissues than in normal tissues. Tumor infiltration by immune cells was confirmed to be correlated with the expression of TMEM115. High TMEM115 expression and immune cell infiltration were further related to poor prognosis in LIHC. We also confirmed a correlation between TMEM115 and TP53 mutations. In conclusion, we confirmed that TMEM115 expression is correlated with tumor-infiltrating immune cells and poor prognosis in LIHC and that TMEM115 shows potential as a prognostic biomarker in LIHC.
Introduction
Liver cancer, the third leading cause of cancer-related deaths worldwide, is expected to affect approximately one million patients by 2025. Liver hepatocellular carcinoma (LIHC) is the most common histological subtype of liver cancer [1]. Over the past decade, strategies for the early diagnosis and treatment of LIHC have improved the survival rate of patients. However, the survival rate of LIHC is low because of the high recurrence, metastasis, and side effects of LIHC chemotherapy [2]. Further, early diagnosis of LIHC remains difficult. Most patients are diagnosed with the disease in the late stages, thus showing poor prognosis after cancer diagnosis. Overall, there is an urgent need for the developing new biomarkers to improve the early diagnosis and survival rate of LIHC; notably, tumor immunotherapy has recently been studied as a new treatment strategy for LIHC.
The immune system plays an important role in cancer progression [3]. Cancer occurs in a complex tumor immune microenvironment (TIME) consisting of immune cells and various extracellular elements. To treat and predict cancer, understanding and overcoming the TIME is essential. The TIME comprises immune cells, such as Th cells, Tc cells, B cells, NK cells, and various other extracellular factors [4]. Tumor-infiltrating immune cells (TIICs) that infiltrate the TIME are related to the prognosis of cancer treatment and have been considered as novel biomarkers that predict therapeutic effects; many relevant studies are also ongoing [5,6]. Therefore, further research on TIICs is important. Many studies have demonstrated a relationship between prognosis and the density and type characteristics of TIICs. These studies have demonstrated the importance of TIICs such as dendritic cells, macrophages, and NK cells in LIHC [7-9].
Transmembrane protein 115 (TMEM115) is a membrane protein that acts as a transporter of proteins from the Golgi to vesicles [10]. Membrane proteins constitute 30% of the human proteins and are used as biomarkers in various physiological and pathological conditions [11]. In addition, Transmembrane proteins have been reported to novel biomarker for prognosis of LIHC [12-14]. However, the TMEM115, a transmembrane protein, has not been confirmed as a biomarker in LIHC; thus, its potential as a tumor biomarker must be confirmed. TP53 mutations occur in many cancers and are associated with poor prognoses [15,16]. Moreover, the most prevalent mutation in LIHC, TP53, has been demonstrated to affect LIHC prognosis [17]. However, the mechanisms underlying the association among TP53 mutations, TMEM115, and TIICs remain to be studied. Therefore, it is necessary to confirm the correlation between TMEM115 and TIICs in the context of TP53 mutations in patients with LIHC.
In this study, we compared the expression of TMEM115 in cancers, including LIHC, with that in normal tissues and evaluated the prognostic value of TMEM115 expression. In addition, we aimed to confirm the correlation between TMEM115 expression and TIICs and to confirm the correlation between TMEM115 expression and TIICs according to TP53 mutations. Thus, we intend to present the possibility of TMEM115 as a biomarker for predicting LIHC prognosis.
Materials and methods
Tumor Immune Estimation Resource (TIMER) database analysis
TIMER (https://cistrome.shinyapps.io/timer/) is an online tool used to analyze gene expression, survival rates, and immune infiltration in various cancers including LIHC; it uses more than 10,000 samples from The Cancer Genome Atlas (TCGA) for analysis [18]. The correlation between TMEM115 and TIICs was analyzed using TIMER. In addition, the correlation between TP53 gene mutations and TIIC was confirmed.
UALCAN database analysis
UALCAN (http://ualcan.path.uab.edu) is an online database that uses TCGA sequences and the clinical data of 31 cancer types [19]. Using UALCAN, we analyzed the differences in gene expression between normal tissues and tumors according to race, sex, histological subtype, age, grade, and stage.
Gene Expression Profiling Interactive Analysis version 2 (GEPIA2)
GEPIA2 (http://gepia.cancer-pku.cn/) is an online database containing information from more than 9000 cancer tissue samples and more than 8000 normal samples [20]. The survival rate and gene expression levels according to gene expression in LIHC were measured using GEPIA2.
OSlihc database analysis
OSlihc was used to evaluate the prognostic value of the gene [21]. The overall survival (OS), decay-free interval (DFI), progression-free interval (PFI), and disorder-specific survival (DSS) related to the gene were measured using OSlihc.
Immunohistochemistry (IHC) staining analysis
The Human Protein Atlas (HPA) is an online database that contains information about protein distribution in human tissues and cells [22]. IHC images were obtained from HPA to confirm the TMEM115 protein expression levels. The protein expression level was expressed as undetected, low, medium, or high according to the fraction of stained cells.
Statistical analysis
The statistical results of all data analyses were automatically analyzed in the online databases, and the statistical results are presented as p-values < 0.05 from a log-rank test and hazard ratio (HR) values.
Results
mRNA expression levels of TMEM115 in LIHC and different tumor types
TMEM115 expression was found to be upregulated in LIHC, bladder urothelial carcinoma, breast invasive carcinoma, bladder cholangiocarcinoma, esophageal carcinoma, head and neck squamous cell carcinoma, and prostate adenocarcinoma compared to normal tissues. In addition, TMEM115 expression was found to be downregulated in colon adenocarcinoma, kidney renal papillary cell carcinoma, lung adenocarcinoma, lung squamous cell carcinoma, thyroid carcinoma, and uterine corpus endometrial carcinoma compared to normal tissues (Fig. 1A). We analyzed the correlation between TMEM115 expression and clinicopathological characteristics, including sample type, race, sex, histological subtype, age, grade, and TP53 mutations in LIHC. Our results showed a significant correlation between TMEM115 expression and the primary tumor, race (Caucasian, African, American, and Asian), sex (male and female), histological subtype (hepatocellular carcinoma, fibrolamellar carcinoma, and hepatocholangiocarcinoma), age, grade (I, II, III, IV), stage (I, II, III, IV), and TP53 mutations in LIHC (Fig. 1B).
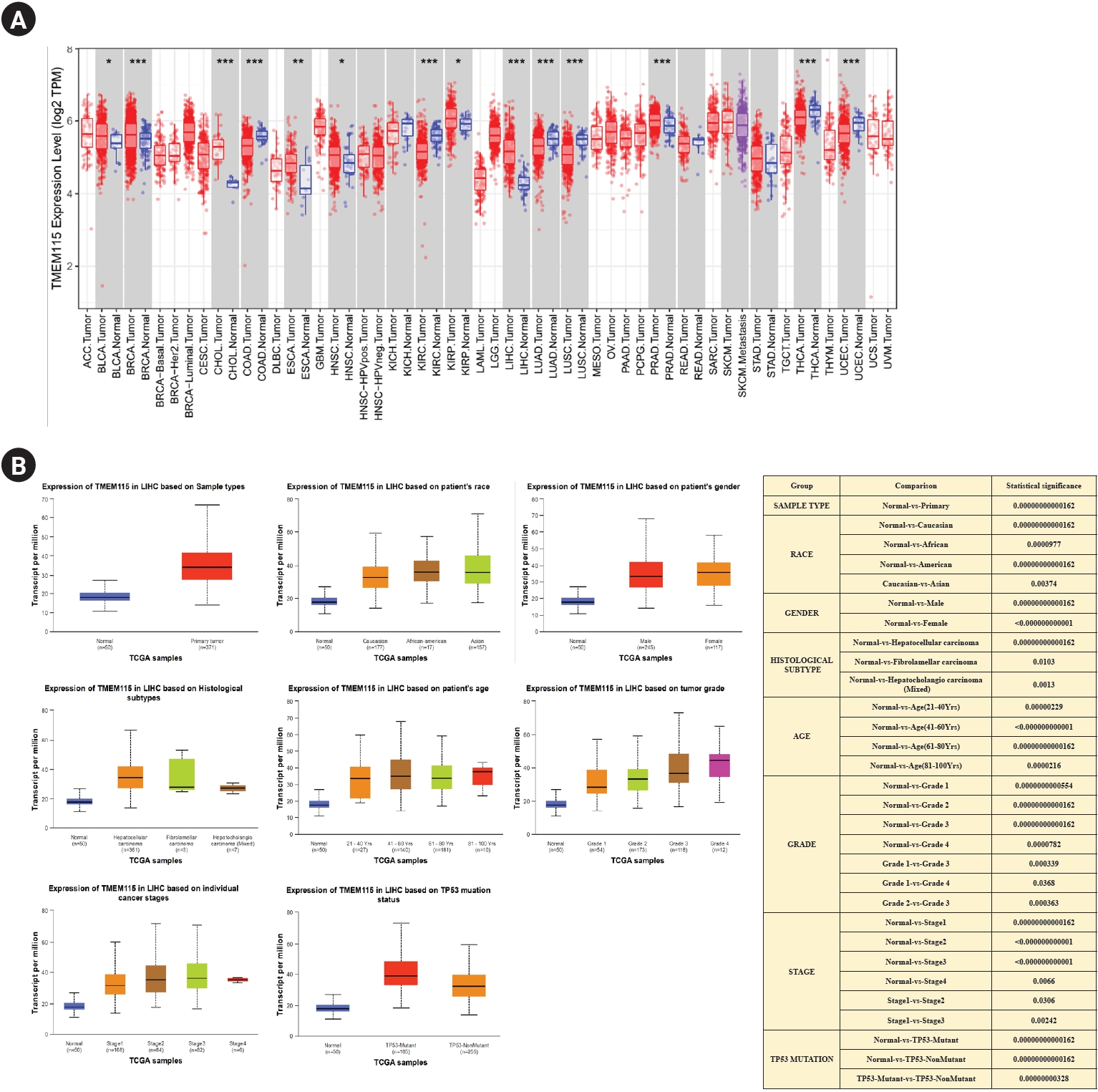
mRNA expression levels of TMEM115
(A) Expression of TMEM115 using the TIMER database, (B) the expression of TMEM115 in various clinicopathologic characteristics using the UALCAN database. TMEM115, transmembrane protein 115; TIMER, tumor immune estimation resource; UALCAN, the university of alabama at birmingham cancer data analysis; ACC, adrenocortical carcinoma; BLCA, bladder urothelial carcinoma; BRCA, breast invasive carcinoma; CESC, cervical squamous cell carcinoma and endocervical adenocarcinoma; CHOL, cholangiocarcinoma; COAD, colon adenocarcinoma; DLBC, diffuse large B-cell lymphoma; ESCA, esophageal carcinoma; GBM, glioblastoma multiforme; HNSC, head and neck squamous cell carcinoma; HPV, human papilloma virus; KICH, kidney chromophobe; KIRC, kidney renal clear cell carcinoma; KIRP, kidney renal papillary cell carcinoma; LAML, acute myeloid leukemia; LGG, lower grade glioma; LIHC, liver hepatocellular carcinoma; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; MESO, Mesothelioma; OV, ovarian serous cystadenocarcinoma; PAAD, pancreatic adenocarcinoma; PCPG, pheochromocytoma and paraganglioma; PRAD, prostate adenocarcinoma; READ, rectum adenocarcinoma; SARC, sarcoma; SKCM, skin cutaneous melanoma; STAD, stomach adenocarcinoma; TGCT, testicular germ cell tumors; THCA, thyroid carcinoma; THYM, thymoma; UCEC, uterine corpus endometrial carcinoma; UCS, uterine carcinosarcoma; UVM, uveal melanoma.
Protein expression levels of TMEM115 in LIHC
The HPA database was used to assess TMEM115 protein expression. IHC results from the HPA showed that TMEM115 protein was not expression in normal liver tissues but was highly expressed in LIHC tissues (Fig. 2).
Prognostic value of TMEM115 expression in LIHC
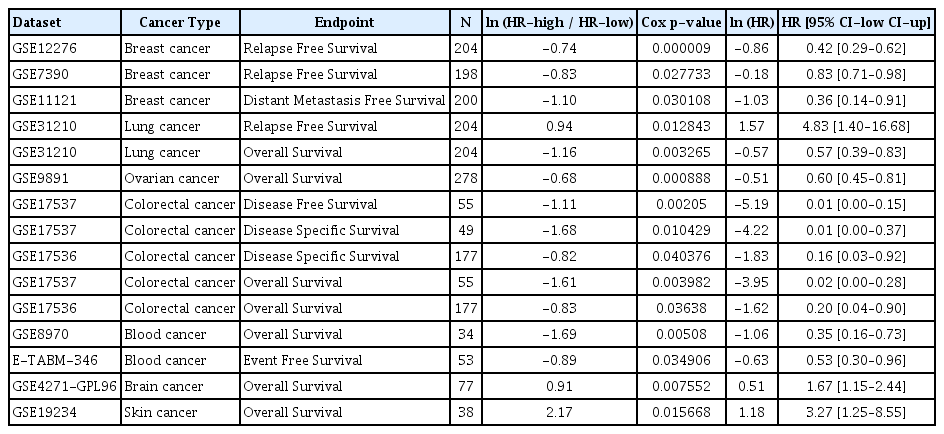
The survival rate for TMEM115 expression in LIHC were analyzed. Higher TMEM115 expression was associated with poorer OS (hazard ratio [HR] = 2, p = 0.00017) and DFS (HR = 1.4, p = 0.04) (Fig. 3A). Further, upregulated TMEM115 expression was correlated with poor prognosis in LIHC using the OSlihc web server (OS: HR = 2.3159, p = 0.00005; DFI: HR = 1.8293, p = 0.0033; PFI: HR = 1.8158, p = 0.0034; DSS: HR = 2.1251, p = 0.0187; Fig. 3B). Furthermore, upregulated TMEM115 expression was associated with poor prognosis in LIHC (age: HR = 1.21, p = 0.0163; sex: HR = 1.21, p = 0.0181; race: HR = 1.23, p = 0.00996; stage: HR = 1.17, p = 0.00669; Fig. 3C). The survival rates associated with TMEM115 expression in various cancers are presented in Table 1.
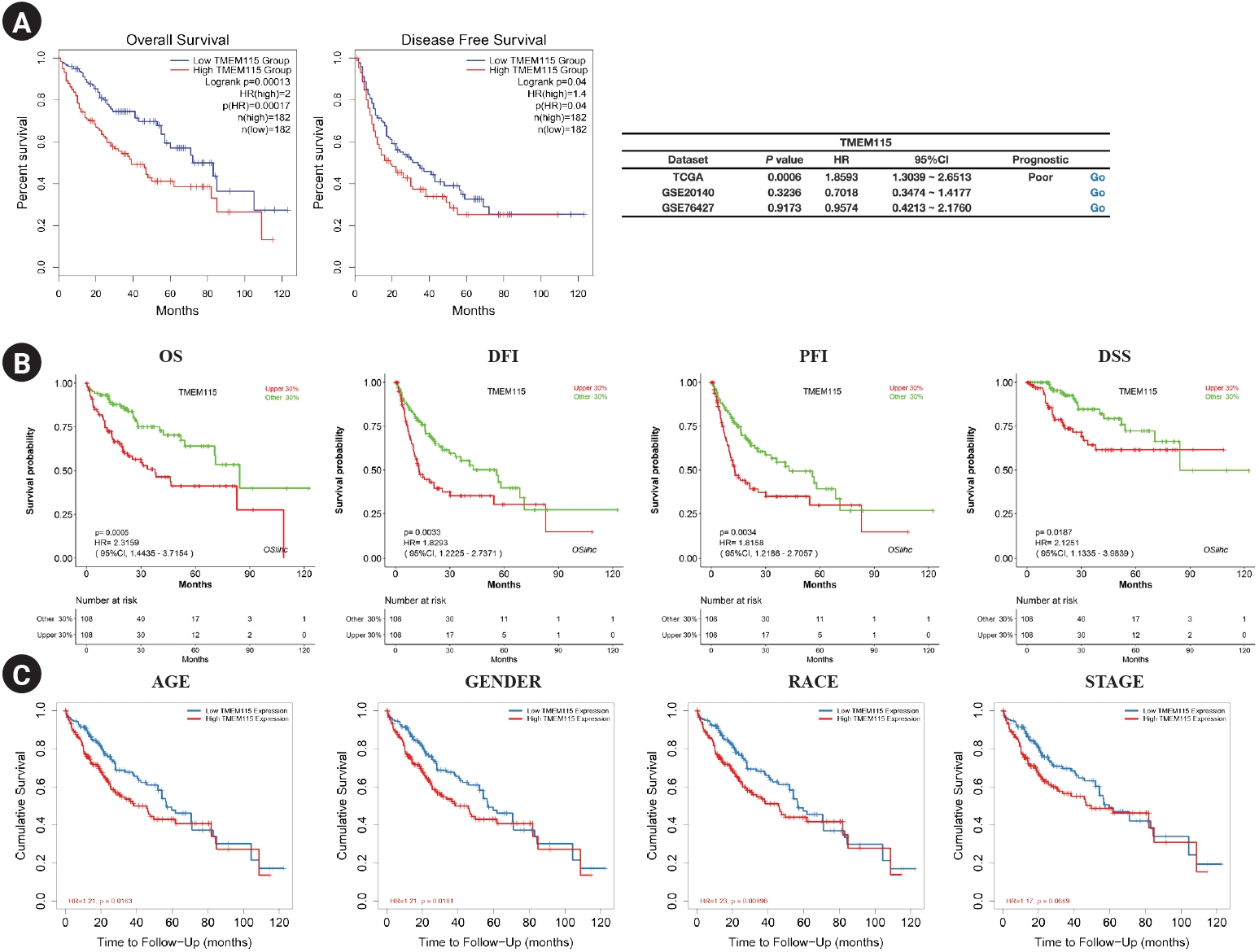
Prognostic significance of TMEM115 expression
(A) GEPIA2, (B) OSlihc, and (C) TIMER. GEPIA2, gene expression profiling interactive analysis 2; OSlihc, online consensus survival web server for liver hepatocellular carcinoma; TIMER, tumor immune estimation resource; TCGA, the cancer genome atlas program; Overall survival, OS; disease-free interval, DFI, progression-free interval, PFI; disease-specific survival, DSS.
Correlations between TMEM115 expression levels and infiltrating immune cells in LIHC
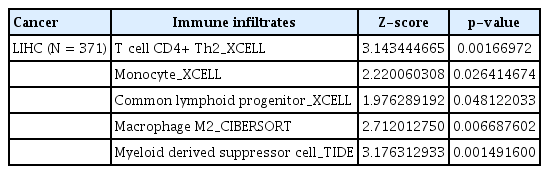
TMEM115 expression levels were positively correlated with the infiltration levels of B cells (r = 0.135, p = 0.012), neutrophils (r = 0.271, p = 0.000000321), macrophages (r = 0.247, p = 0.00000358), myeloid dendritic cells (r = 0.277, p = 0.00000017), CD4+ T cells (r = 0.105, p = 0.022), and CD8+ T cells (r = 0.161, p = 0.0028; Figure 4A). In addition, TMEM115 expression levels were positively correlated with the infiltration levels of follicular helper T cells (r = 0.16, p = 0.0027), regulatory T cells (r = 0.11, p = 0.033) and M0 macrophages (r = 0.12, p = 0.02), whereas resting CD4 memory T cells (r = -0.14, p = 0.0066) and resting mast cells (r = -0.12, p = 0.019) were negatively associated with TMEM115 expression levels (Fig. 4B). We investigated whether TMEM115 expression was associated with prognosis and TIICs in LIHC. High TMEM115 expression and high neutrophil infiltration levels were associated with worse prognosis than that with low TMEM115 expression and low neutrophil infiltration levels. High TMEM115 expression and high macrophage infiltration levels were associated with a worse prognosis than that with low TMEM115 expression and low macrophage infiltration levels. High TMEM115 expression and high CD4+T cell infiltration levels were associated with worse prognosis than that with low TMEM115 expression and high CD4+T cell infiltration levels (Fig. 4C). In addition, based on the XCELL algorithm, the infiltration level of T cell CD4+ Th2, monocyte, common lymphoid progenitor was positively associated with the TMEM115 expression. Based on the CIBERSORT, the infiltration level of macrophage M2 was positively associated with the TMEM115 expression. Based on the TIDE, the infiltration level of myeloid derived suppressor cell was positively associated with the TMEM115 expression (Table 2). Taken together, our results suggest that High TMEM115 expression is associated with TIICs and may affect tumor prognosis.
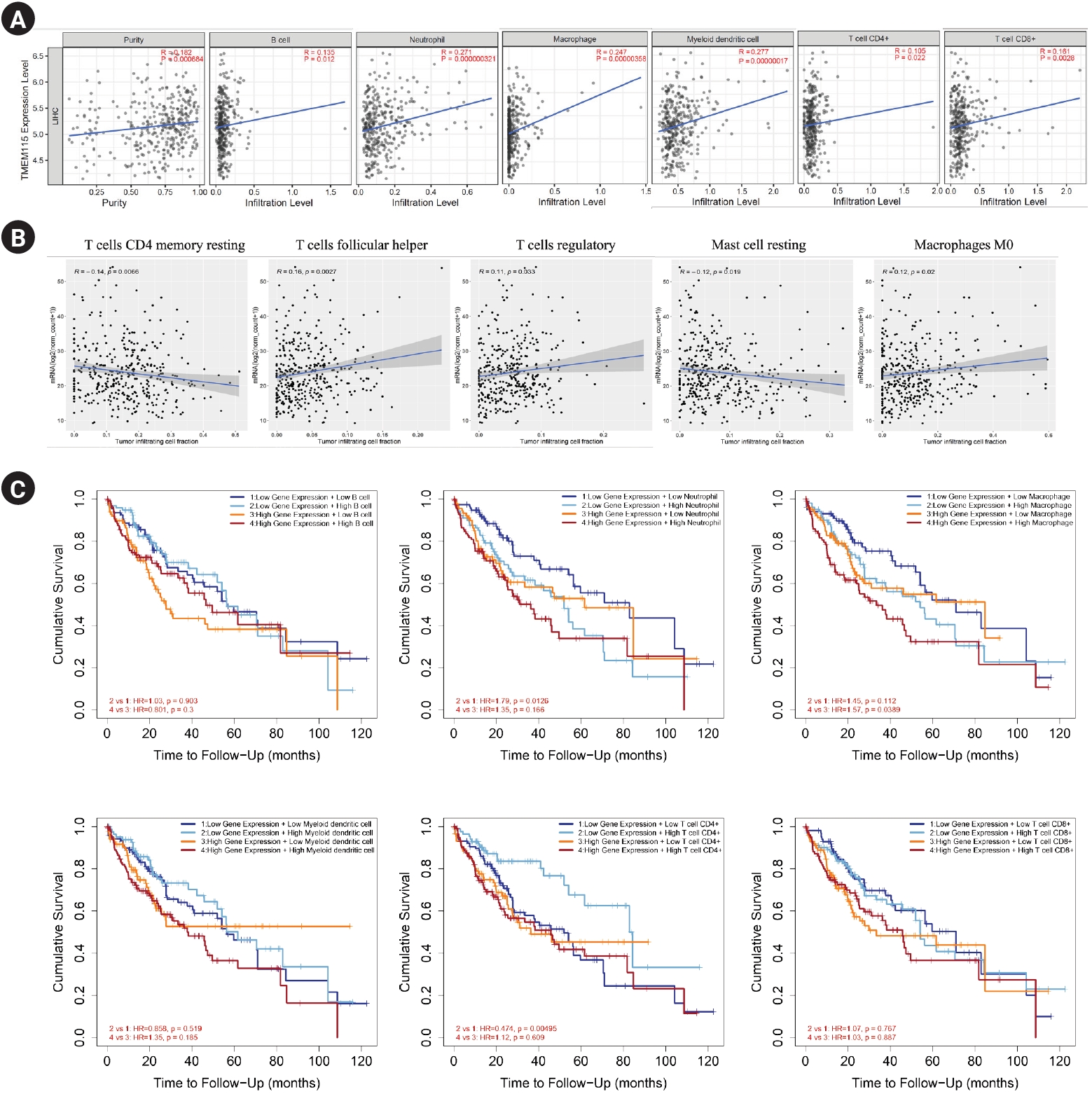
Correlation between TMEM115 expression and TIICs
. (A) The correlation between TMEM115 and TIICs, (B) the correlation between the expression of TMEM115 and immune cells, (C) the prognostic value between TMEM115 expression and TIICs. TMEM115, transmembrane protein 115; TIICs, Tumor-infiltrating immune cells; CD4, cluster of differentiation 4; CD8, cluster of differentiation 8.
Correlation between TMEM115 expression and TP53 mutation in LIHC
The correlation between TP53 mutation and TMEM115 expression in LIHC was investigated. TMEM115 expression levels were positively correlated with the TP53 expression levels (p = 0.0000174) (Fig. 5A). In addition, TMEM115 expression was increased in TP53 mutation compared to that in the wild type (WT) (Fig. 5B). TMEM115 mutation was increased the number of macrophages M2 and activated memory CD4+ T cells (Figure 5C). Further, TP53 mutation was associated with higher infiltration levels of B cells and macrophages (Fig. 5D). Taken together, these results suggest that TMEM115 expression is associated with TP53 mutations.
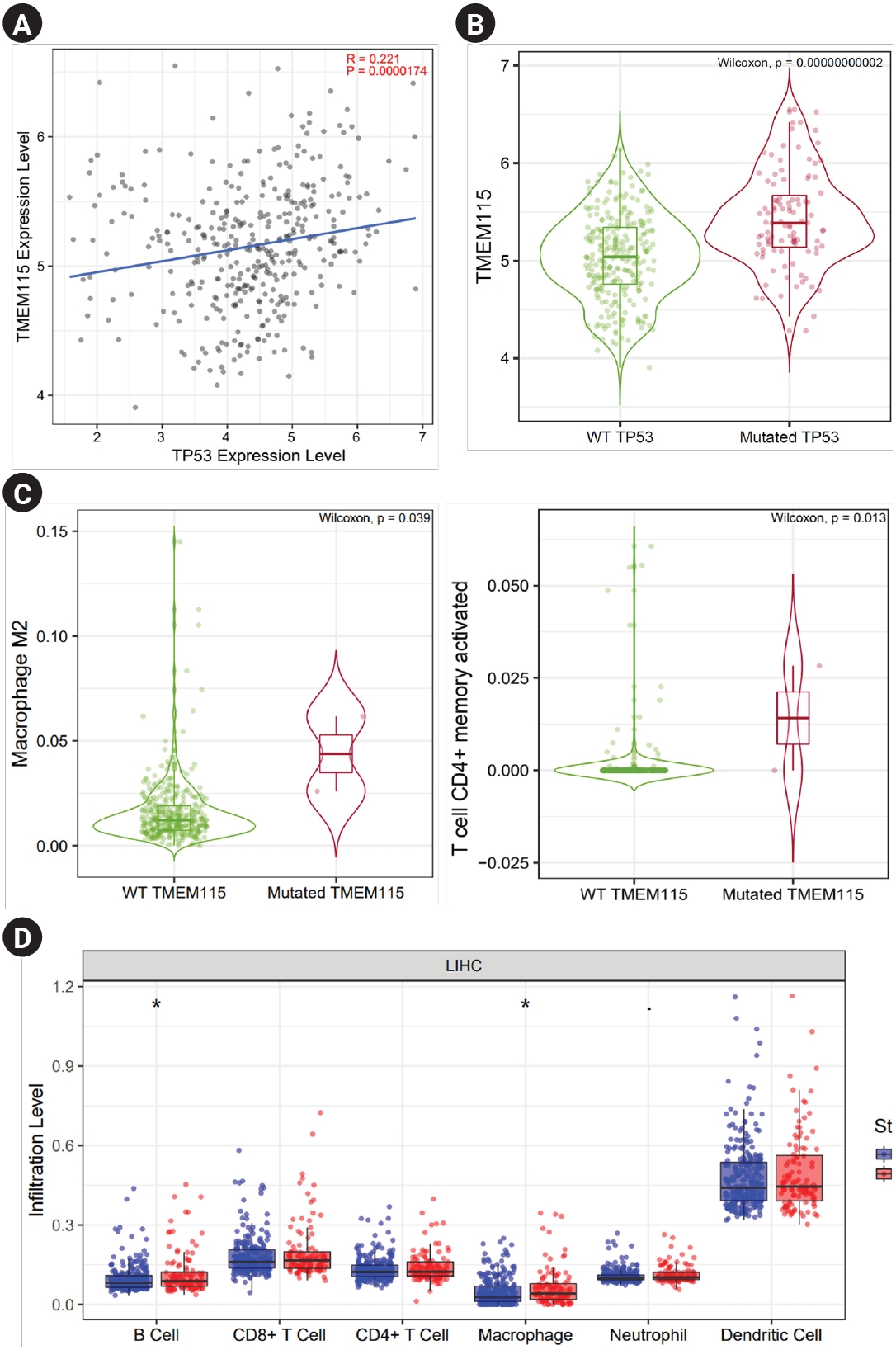
Correlation between TMEM115 expression and TP 53 gene mutations
(A) The correlation between TMEM115 expression and TP53 gene expression, (B) the correlation between TMEM115 expression and TP53 gene mutations, (C) the correlations between tumor-infiltrating immune cells and TMEM115 mutation, (D) the correlation between TP53 expression and TIICs in LIHC. TMEM115, transmembrane protein 115; WT, wild type; LIHC, liver hepatocellular carcinoma.
Discussion
Among various cancers, liver cancer has a high mortality rate worldwide [23]. LIHC is the most common pathological form of liver cancer and occurs through chronic liver inflammation [24]. LIHC is associated with a continuous increase in mortality owing to its poor prognosis and limited treatment methods [25]. Biomarkers for prognostic prediction and treatment strategies for LIHC are still being researched, but novel biomarkers have not settled in clinical practice except for alpha-fetoprotein [26]. Therefore, research is being performed to create biomarkers in order to uncover novel treatment techniques for LIHC.
The immune system plays an important role in the progression and growth of cancers. In the TIME, TIICs surrounding tumor cells play an important role in cancer growth and have been found to be involved in the development and progression of cancer; they are thus being studied as important prognostic indicators and potential therapeutic targets [27,28].
TP53 is a tumor suppressor gene involved in apoptosis, cell cycle arrest, and DNA repair. However, mutations in TP53 are known to cause abnormal cell proliferation and tumor gene activity, which are associated with poor prognosis in cancer. Research on TP53 mutations and immune cell regulation is constantly being conducted to study tumor immune regulation [29,30]. However, the mechanisms by which TP53 mutations are related to TMEM115 expression and TIICs have not been studied.
TMEM115 is a membrane protein; notably, membrane proteins constitute 30% of human proteins and have been used as a physiological and pathological biomarker [10,11]. Fan et al. found that increased TMEM147 expression was associated with poor prognosis [14]. Furthermore, LIHC has demonstrated the potential of transmembrane protein as a biomarker. In this study, the mRNA level of TMEM115 was found to be higher in LIHC tissues than in normal tissues. In addition, by upon analyzing the gene expression level by race, sex, age, grade, and stage in detail, a high expression level of TMEM115 was confirmed compared to that in normal tissues. TMEM115 expression was also correlated with poor prognosis in LIHC. In addition, a correlation between poor prognosis and various clinicopathological factors was confirmed. Thus, TMEM115 may serve as a potential biomarker for LIHC.
Protein expression is important for gene function. By comparing the protein expression levels of TMEM115 in LIHC tissues, IHC analysis confirmed its higher expression in LIHC tissues than in normal tissues. These results suggest an effect of TMEM115 at the transcriptional and translational levels in patients with LIHC.
We confirmed the correlation between TMEM115 expression and immune cell infiltration in LIHC. High TMEM115 expression and immune cell infiltration was found to be associated with worse prognosis in LIHC. These findings were consistent with previous research on the identification of LIHC biomarkers [31,32].
TP53 mutation, which is key to the progression of malignant tumors, results in loss of function through genetic modification in more than half of human cancers [33,34]. Therefore, the correlation between TP53 and TMEM115 in the context of LIHC prognosis, needs to be confirmed. In a correlation investigation between TP53 mutation and TMEM115 expression in LIHC cells, TMEM115 expression was increased in TP53 mutation compared to that in WT. This finding suggests that TMEM115 is associated with TP53 mutation.
In conclusion, we suggest that high TMEM115 expression correlates with poor prognosis in LIHC. Therefore, we present TMEM115 as a potential biomarker for LIHC. As the results of this study were confirmed using online databases, further studies are needed to confirm the potential function of TMEM115 using in vitro and vivo models.
Notes
Conflict of interest
The authors declare no conflicts-of-interest related to this article.