 |
 |
- Search
| Keimyung Med J > Volume 39(1); 2020 > Article |
|
Abstract
Acknowledgements
Notes
Funding
The present study was supported by the National Research Foundation of Korea (grant funded by the Korea Government Ministry of Science, ICT and Future Planning; grant nos. 2014R1A5A2010008, NRF-2016R1D1A1B04930619, and NRF-2019R1I1A3A01063114).
Availability of data and materials
The datasets analyzed during the present study are available from The Cancer Genome Atlas (https://www.cancer.gov/tcga) and the UCSC Xena (https://xena.ucsc.edu). The datasets generated in the present study are available from the corresponding author upon reasonable request.
Authors contributions
WJP and SK contributed to the conception and design of the study, analysis of the data, interpretation of results, and the writing of the manuscript. JYP and SK contributed to the acquisition of data and the writing of the manuscript. TKK, JWP, and SK reviewed the manuscript. SK edited the manuscript. All authors read and approved the manuscript and agree to be accountable for all aspects of the research in ensuring that the accuracy and integrity of any part of the work are appropriately investigated and resolved.
Fig. 1.
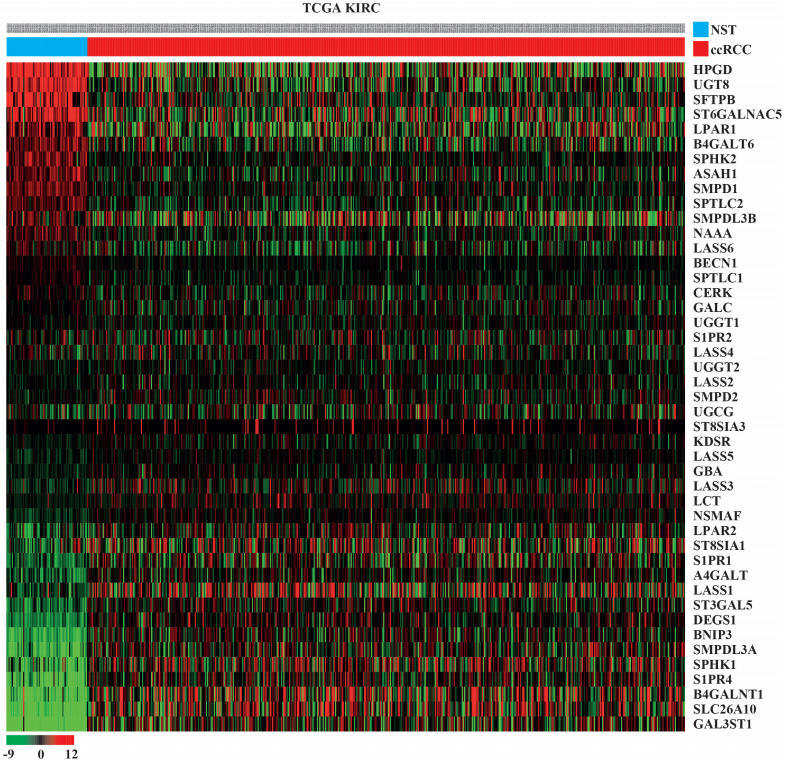
Fig. 2.
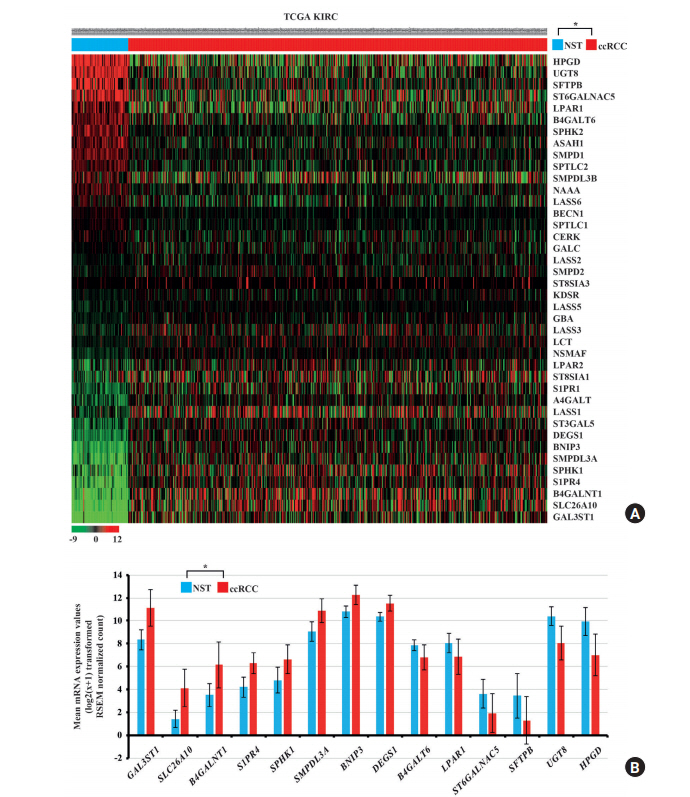
Fig. 3.
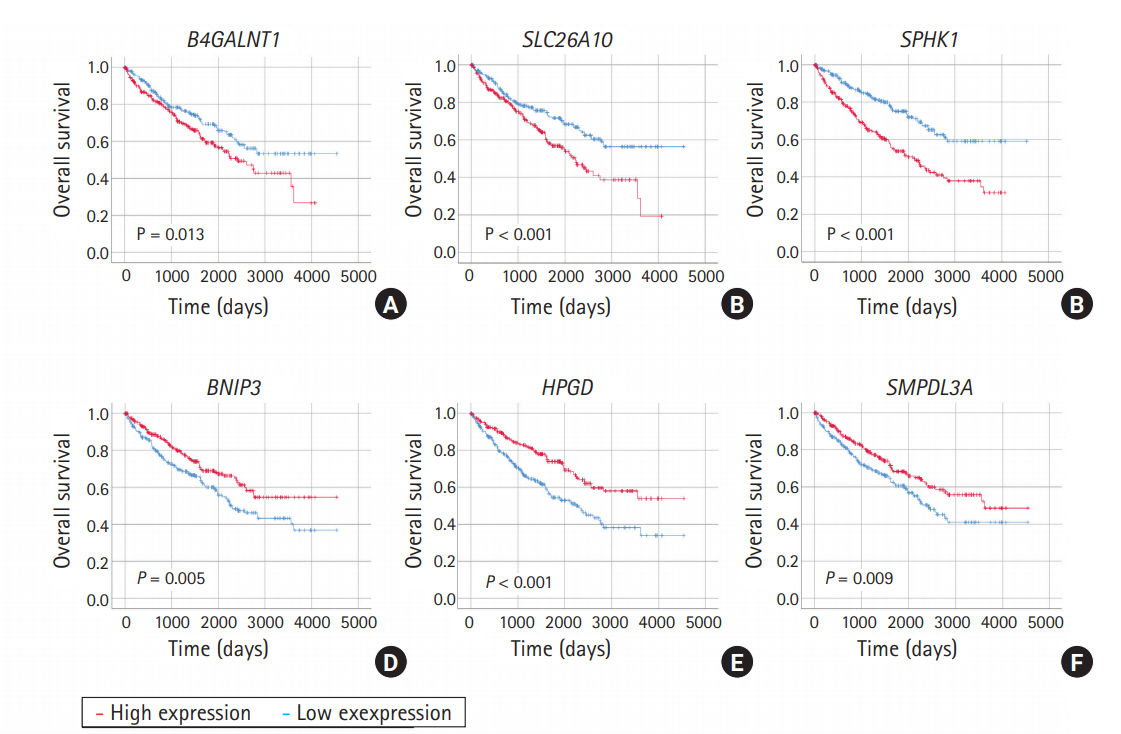
Table 1.
| Samples | Correlations between components | Pearson’s correlation coefficient value | p-Value* |
|---|---|---|---|
| TCGA KIRC tissues from TCGA KIRC cohort (n = 533) | B4GALNT1 and B4GALT6 | -0.271 | < 0.001 |
| B4GALNT1 and BNIP3 | -0.129 | 0.003 | |
| B4GALNT1 and GAL3ST1 | -0.267 | < 0.001 | |
| B4GALNT1 and SLC26A10 | 0.768 | < 0.001 | |
| B4GALNT1 and SMPDL3A | -0.244 | < 0.001 | |
| B4GALNT1 and SPHK1 | 0.181 | < 0.001 | |
| B4GALNT1 and ST6GALNAC5 | 0.087 | 0.045 | |
| B4GALNT1 and UGT8 | -0.110 | 0.011 | |
| B4GALT6 and DEGS1 | -0.141 | 0.001 | |
| B4GALT6 and GAL3ST1 | -0.127 | 0.003 | |
| B4GALT6 and HPGD | 0.158 | < 0.001 | |
| B4GALT6 and S1PR4 | -0.232 | < 0.001 | |
| B4GALT6 and SFTPB | 0.352 | < 0.001 | |
| B4GALT6 and SLC26A10 | -0.346 | < 0.001 | |
| B4GALT6 and SMPDL3A | 0.120 | 0.006 | |
| B4GALT6 and SPHK1 | -0.255 | < 0.001 | |
| B4GALT6 and ST6GALNAC5 | -0.183 | < 0.001 | |
| B4GALT6 and UGT8 | 0.347 | < 0.001 | |
| BNIP3 and DEGS1 | 0.123 | 0.004 | |
| BNIP3 and GAL3ST1 | 0.410 | < 0.001 | |
| BNIP3 and HPGD | -0.106 | 0.014 | |
| BNIP3 and LPAR1 | -0.201 | < 0.001 | |
| BNIP3 and SFTPB | -0.353 | < 0.001 | |
| BNIP3 and SLC26A10 | -0.104 | 0.016 | |
| BNIP3 and SMPDL3A | 0.419 | < 0.001 | |
| BNIP3 and SPHK1 | -0.320 | < 0.001 |
References
-
METRICS

-
- 2 Crossref
- 4,413 View
- 81 Download
- Related articles in Keimyung Med J





